Data Based Congestive Heart Failure Predictions
The skills the authors demonstrated here can be learned through taking Data Science with Machine Learning bootcamp with NYC Data Science Academy.
Congestive Heart Failure (CHF) is one of the most prevalent diseases in the United States, contributing to ~10% of patient admissions. In this project, we aim to ease this burden and provide a valuable tool for clinicians and patients utilizing the power of data. The data set we used was the Medical Information Mart for Intensive Care (MIMIC- IV) which comprised of 260,000 patients and 500,000,000 events recorded for those patients' hospital admissions.
We manually cleaned the data by correcting errors that would arise when converting lab results from string to numeric types and imputed missing values using KNN and normal value imputation. Our models' thresholds were adjusted to have 95% recall because the cost of a false negative is potential death of a patient while that of a false positive is additional testing. We trained logistic regression, random forest and XG boost models to predict current and future diagnosis of CHF.
Most of our models had a precision of ~25% when adjusting for 95% recall. These models can be used as a decision support tool to flag patients for additional testing in a clinical setting and an early warning tool to educate patients to make lifestyle changes.
Introduction to Healthcare Data and CHF
In today’s digitized network of healthcare data where we collect more patient data than ever before, there is an increase in demand for analytics tools to guide clinicians in their daily workflows. Healthcare organizations are even incentivized to collect patient data electronically and improve patient outcomes through the government sponsored Meaningful Use program, leading to a great increase in the volume and utilization of patient health data. As data scientists, we aim to utilize this wealth of information available to make a positive impact on the patient population.
For this project, we chose to investigate Congestive Heart Failure (CHF). CHF is a highly prevalent disease that affects 6 million people in the US and contributed to 13% of total deaths in the US in 2012, costing the nation over $30 billion annually. CHF is also a deadly disease with ~50% 5-year survival rate. The severity of CHF is broken down into stages A~D with stage D being the most severe.
One study conducted by the American Heart Association showed that the 5-year survival rate dropped 20% as patients progress from Stage B to Stage C. This shows that we can greatly increase the survival rate in patients if we can detect CHF in patients earlier and prevent them from progressing to a more severe form.
Data Source
The data that we used for this project is from the Medical Information Mart for Intensive Care (MIMIC-IV). This version of the data set was introduced in 2020 and is composed of patients from Intensive Care Units and Emergency Departments from Beth Israel Deaconess Medical Center in Boston, MA from 2008~2019. In order to gain access to this data set, we needed to pass an ethics course that details the importance of anonymity in patient data to ensure that the data is used to bring value to patients rather than maximizing a company’s profits.
MIMIC-IV data set contains a total of 34 tables of which we used 8 tables shown in the diagram below. There was a total of 260,000 patients and 200,000 adult patients recorded in our data set. These patients contributed to a total of 530,000 admissions and 460,000 adult admissions. From these admissions there were over 500,000,000 events including lab results for all admissions and vital information for just ICU admissions.

Data Processing - Pt. 1
Our data set contained hundreds of thousands of patients with hundreds of millions of data points. To make our tasks and modeling more manageable we decided to work with just a few samples of the population, some for current predictions and one for future predictions. We found that approximately 10.7% of all hospital visits resulted in a diagnosis of heart failure, which matched what Brandon had just talked about, so we ensured that our samples retained the same ratio.
We also made sure, for our positive diagnosis patients, to use the first hospitalization in which they received the diagnosis and for our negative patients that they never had any hospitalization in which they received a positive diagnosis for heart failure. For positive patients this helped us to train our models to catch initial cases. This is important in trying to combat the issue as early as possible. For negative patients we didn’t want to confuse our model with labs that might be taken very shortly before a positive diagnosis would be made.
Similar to positive diagnosis, we also checked to make sure that our age, gender and ethnicity distributions matched between our samples and full data.
Data Processing - Pt. 2
After we had these sample sets of patients, we had to trim some due to age. We knew age would be an important factor, but due to anonymization of the data it was impossible to determine anything for patients under 18 years old, so we dropped them. We also lost a number of patients who had no laboratory results. We can’t say if this is because they actually didn’t have any tests done at the hospital or if because it simply wasn’t included in the data set. All told this brought our current predictive sample down to about 11 and a half thousand and our future predictive sample to about 8 thousand.
Feature Engineering - Pt. 1
The largest set of data we have to help make our diagnoses is lab results. The Lab results table houses over 120 million records which cover 1625 different tests (though there are likely some overlaps in these types of tests). That said, not every patient takes every test during each hospital visit. The vast majority of these tests would present missing data for any given hospitalization. On the flip side, there are a number of tests which are taken repeatedly and at regular intervals during a single hospitalization.
To try and get the most relevant lab results we decided to grab the most commonly run labs on our sample set. This should help mirror how our model might run in an actual hospital setting on those tests most common to incoming patients. Separately we did some outside research to see if there were any labs commonly related to Congestive Heart Failure diagnosis. After grabbing the top 20 commonly run labs and 5 outside research labs we ran some aggregation to account for the multiple lab results.
Each lab had expected lower and upper bounds given for a healthy range. These values could be slightly different depending on gender and ethnicity. We calculated the min, mean and maximum value for each patient, hospitalization and lab ID combination. We also counted the number of labs which were outside of the expected range and divided by the total number of labs taken to get an Abnormal percent. Finally, for any patient and hospitalization where a lab had abnormal values, we kept the highest value above the healthy upper bound and lowest value below the healthy lower bound.
This gave us 150 total lab event related features.
Feature Engineering - Pt. 2
To this list of features, we added some general patient statistics. The first we had to engineer, co-morbidities, which are other prior diagnoses which might lead to congestive heart failure. We mapped our sample patients onto prior hospitalization records within 3 years of their current diagnoses in order to grab this information. We chose 3 years so these prior diagnoses could reasonably be considered related to current health situations. For our future prediction model, we only used a 1-year difference as we wanted to ensure the related labs were relatively recent for the heart failure diagnosis.
Beyond this we added emergency room stay duration, age, gender, ethnicity, and whether the patients had medicare or medicaid. Features which we saw, or thought would be likely to have an impact on a positive congestive heart failure diagnosis. There was a lot of data cleaning needed, parsing text comments for numeric values, removing double periods, mapping text such as Negative, Trace, Small, Moderate, Large; things like this.
When that was done and the features which needed it were standardized, we were left with 173 features and lots of multicollinearity. After running a recursive check for variance inflation factor, we were able to reduce this set to around 110 features depending on the sample.
Imputation
Our next problem to deal with was missingness. For many of the labs, especially the commonly performed ones, we thought the data was Missing At Random. We think the reason the lab wasn’t taken was because the doctor didn’t need it, they either assumed it was normal or they could infer the value based on other lab results. This led us to two different methods of imputation.
For the first we created a normal distribution of 50 randomly generated values around the middle of the healthy range. This would give us a very small chance at returning an abnormal value. We also created a second set of data imputing with K nearest neighbors. This would be more likely to return abnormal values depending on the existing data of other patients.
As we learned later both of these methods returned very similar results.
Data Driven Feature Selection
With our missingness no longer an issue we still wanted to address our model complexity, the number of features. We first tried Principal Component Analysis which reduced our feature set to 35 without a large reduction in Recall or Area Under the Curve, our primary target. You’ll see the recommendation was slightly higher, but we found no great loss in using only 35 features. However, this obviously made our model harder to interpret and as we found later didn’t provide significant benefits to our scoring metrics.
Instead, we focused on Recursive Feature Elimination. For current predictions recursive elimination reduced our feature count to 36 for logistic regression, 60 for XGBoost, and only 101 for random forest. This was without reducing any interpretability, in fact making them less complex, for the most part.
For our future prediction models feature selection was done more manually and resulted in 78 features for each model.
Final Samples for Models
After all this we ended up with four separate sample populations, one for current predictions with all types of patients, one each for current predictions split by Male and Female, and one for future predictions. The total number of patients in each were on the same scale, 11 and a half thousand for current full and 8 to 8.7 thousand for the rest.
Though the Male sample had a larger percentage of positive diagnosis than the rest, it was still in line with our general male population above the age of 18. We think this increased likelihood is likely due to males having more heart issues in general.
For our logistic model for current predictions, we were able to use only 36 features, while our remaining models all had around 60-80 features, with the exception of Random Forest at 101.
Unsupervised Modeling
Our lab tests had over 100 independent variables, so we used an unsupervised model to better understand the feature space. We chose Principal Component Analysis (PCA), because PCA is good for reducing a large feature space of continuous data.
In the PCA, we found 35 components were sufficient to explain the majority of the variance of the overall 100 independent variables. When these components were applied to a logistic regression, they achieved 80% accuracy on a balanced test set. The relatively high accuracy of the PCA with only 1/3 of the data confirmed our hypothesis that only a fraction of the lab tests were providing useful information.

Supervised Modeling
Our team predicted congestive heart failure in three ways:
A group of models to...
- Compare the different risk factors of CHF for male vs female.
- Predict whether someone currently has CHF.
- Predict whether someone will get CHF within one year.
When tuning the model parameters, we optimized for the receiver operating characteristic, also known as the area under the curve (ROC/AUC). In an unbalanced problem like CHF where only 12% of the patients are positive, AUC is better than accuracy for optimization, because the AUC measures the fraction of correctly identified subjects instead of the absolute number. After tuning the model for AUC, we then adjusted the classifier threshold for a 95% sensitivity, which meant that the model will only miss 5% of the actual CHF patients.
Model 1 - Comparing Males and Females
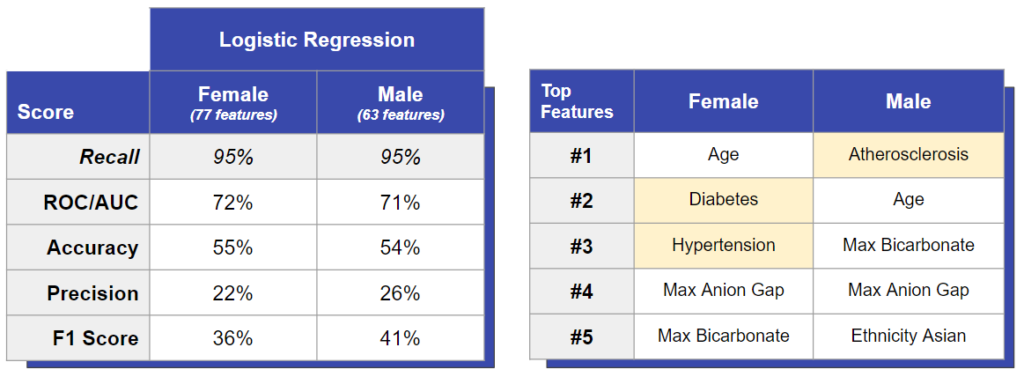
We used two logistic classifiers to predict CHF in males vs females. One model was trained and tested using only male patients, and the other was trained and tested using only female patients. We saw that both models performed similarly in terms of accuracy and precision, which tells us that CHF is not easier or harder to diagnose in males or females. However, we saw important differences in the feature importances between the two.
For example, even though females have lower rates of diabetes and hypertension, we saw these diseases caused a great risk for CHF in females. This suggests that diabetes and hypertension are more damaging to female hearts. On the other hand, males have higher rates of atherosclerosis and we saw atherosclerosis as the biggest risk factor for males, which may explain why men overall have higher rates of CHF than women.
Model 2 - Present (Supervised)
We used three models to predict whether someone currently has CHF. First, we chose a logistic classifier, because this operates most similarly to how doctors classify patients as sick based on high or low values. We also tried random forest classifiers and gradient tree boosting classifiers, because these models are best at finding hidden patterns in the data. When training all three, we used standardization and under/over-sampling to find the best model for AUC.
As typical with data science competitions, we saw the best performance with the gradient boosted model (XGBoost). However, the XGBoost model only performed slightly better than the logistic model. When comparing feature importances, the gradient boosting model relied most heavily on different lab tests, while the logistic model relied most heavily on comorbidities and only a handful of lab tests. In this case, the increased accuracy of XGBoost may not be worth the increase in model complexity compared to the interpretability of logistic regression.

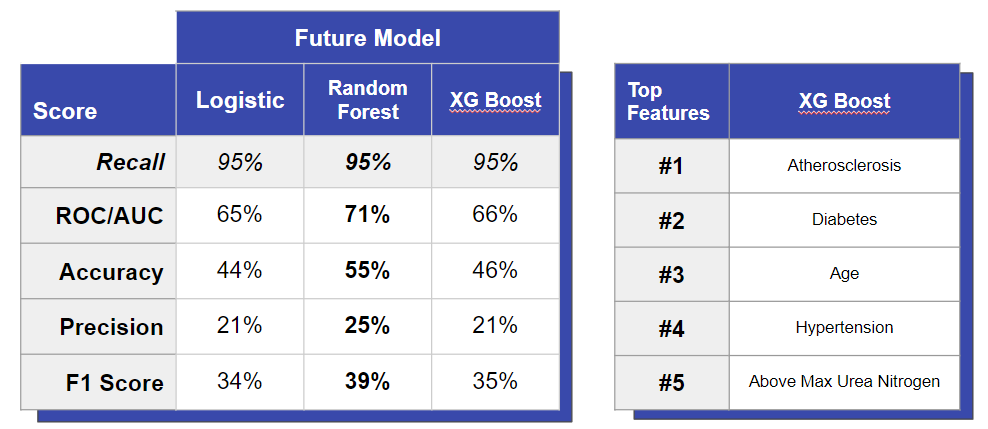
Model 3 - Future (Supervised)
We used the same three types of models as above to predict whether someone will get CHF within one year. This was done by identifying hospital visits right before a patient came in with CHF - when they were still negative - and training a model to classify the pre-heart failure condition.
In this case, we saw the best performance with the random forest (not XGBoost). When comparing feature importances, the random forest was the best at identifying how comorbidities like diabetes and hypertension increase the risk for CHF. In addition, saw the logistic model performing similarly well to the random forest. Once again, this shows the robustness of logistic regression compared to more complicated models.
Data Based Conclusion
Through this project, we had the opportunity to overcome some difficult challenges to create valuable insights for the patients. The sparsity of the dataset allowed us to utilize imputation methods such as K-Nearest Neighbors and normal value imputation. We also cleaned our dataset manually by resolving errors when converting lab results to numerical values. As a result of our modeling, we found that our present model acted in a similar way that doctors clinically diagnose patients by looking at maximum and minimum values of lab results.
We also found that the most important comorbidity for females was diabetes while that for males was atherosclerosis. Our future model had a similar accuracy as our present model but relied more heavily on comorbidities rather than lab results. These models can be used as a decision support tools to screen for patients who may be at risk or use as an early warning tool for patients to make an active choice in changing their lifestyles.
Future Work on Healthcare Data
There are more opportunities that we would like to pursue but were limited with the timeline of the project. As we did with separating genders, we can try separating ethnicities and compare accuracy and top features. We can also try to ensemble different types of models using the Sklearn voting classifier to see if we can improve our precision scores. Another approach to increasing our precision would be to expand our feature set by incorporating more tables such as the prescription table. Finally, we can classify patients at risk of dying from CHF within a year to predict the severity of the condition.
References
The Github repo for all the code in this project can be found here.
This project was inspired by and took direction from a previous capstone project by Eric Myers, Haoyun Zhang, and Maite Herrero Gorostiza. Their work can be found here.
The open-source hospital data was made possible by Johnson, A., Bulgarelli, L., Pollard, T., Horng, S., Celi, L. A., & Mark, R. (2020). MIMIC-IV (version 0.4). PhysioNet. https://doi.org/10.13026/a3wn-hq05.
Access to the data was provided by Goldberger, A., Amaral, L., Glass, L., Hausdorff, J., Ivanov, P. C., Mark, R., ... & Stanley, H. E. (2000). PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation [Online]. 101 (23), pp. e215–e220.

