Detecting Thyroid Lesions in Ultrasound Scans Using Deep Learning
Introduction
My group and I partnered with the NYC office of Koios Medical (Koios website) to locate lesions in ultrasound scans. Up until this point, radiologists have “eye-balled” ultrasound scans to determine whether or not the lesion is malignant or benign. However, this can be problematic because it is, unfortunately, common for doctors to incorrectly classify ultrasound scans, which can put unnecessary tremendous stress on patients if given a false diagnosis. Machine learning techniques can be used to correctly classify medical scans, which will ultimately make the process more efficient and easier for patients.
Data & Methodology
This study uses deep learning technologies to locate lesions in ultrasound scans, so that the correct diagnosis can be delivered to patients. The data was obtained from a public source called Cimalab which is a research facility in Colombia. Three datasets (available here) were publicly available that included characteristics of the patient, coordinates of the bounding box within the image, and coordinates of the lesion. Bounding boxes refers to rectangles that locate objects in the image. Segmentation, on the other hand, refer to the polygons of the actual lesions which have more points than a bounding box. Half of our group implemented segmentation and the other used the bounding box method. I used the bounding box method which I will discuss further in detail.
I had two main variables of focus: the local file paths of the ultrasound images and the bounding box coordinates. The ultrasound images were raw .JPG files that will be eventually transformed into mathematical arrays and fed into the model. Bounding box coordinates refer to the x,y coordinates of the top left corner, the width, and the height. The width and height were added to the x,y coordinates to represent the bottom-right corner. These two sets of coordinates that represented opposite rectangle corners were going to be the values that the model predicts in this regression problem.
In a more traditional machine learning problem, the first few steps would be to fill in missing values and conduct feature engineering. However, because the only inputs to the model were the raw images, this was not necessary. Other adjustments were needed to prepare the inputs and the outputs to the Keras neural network.
The dataset included 488 observations and images were augmented to increase the number of cases to almost 3,000. Four changes were made to the original images: horizontal mirror, rotation, brightness, and contrast. The model sees these transformed images as separate images and it is an effective method to deal with data scarcity. The training dataset contained 60% of the cases, the validation set contained 20%, and the test set contained 20%.
Convert images to arrays
The first way we converted raw images into numpy arrays was by using the keras preprocessing built-in functions. The dimensions for all images were set to 128 x 128, which was reccomended to us by Koios. This dimension size is relatively small, which is optimal for computational processing, but large enough to retain important features in the image. In order to resize the image, however, we needed to adjust the coordinates accordingly because once the image is adjusted, the coordinates would no longer corresponding to the correct location in the ultrasound scan. Thus, whatever numeric transformation was applied to the image dimensions, was also applied to its set of bounding box coordinates. After these transformations, the images were in the correct format to be fed into the model.
The second method we converted raw images into arrays was with Keras image generators, which automatically takes the raw images as an input and outputs neural network tensors, which are just mathematical arrays. This process is undoubtedly simple than the former, but Image Generators made it difficult for me to understand the underlying processes, which is why I chose to explore routes around the Keras generators.
Convolutional Neural Network
Koios Medical, our corporate partner, suggested that we use a neural network implemented with Keras, which is an open-source library used in Python. The neural network takes arrays as inputs, which meant that we would have to transform the ultrasound scans into arrays. For our research problem, our model would output the four bounding box coordinates, which was convenient because they were already structured in a four dimensional array.
The type model used for this analysis was called a convolutional neural network (CNN), which is commonly applied to object detection tasks like this one. CNN’s are able to effectively detect and learn characteristics from images, which is extremely useful if require to make predictions on new images. CNN’s historically generate better predictions that traditionally neural networks.
Additionally, high-performing pretrained models can be loaded from Keras and used to solve other image detection problems. These models have pretrained weights that can be adjusted once they are applied to new problems. This study used a pretrained model called Xception which was trained on a different dataset called “ImageNet”, which has information about objects in outdoor environments as opposed to ultrasound scans. These two setting obviously differ drastically, but transfer learning is known to give great results regardless of those differences.
Four main layers were used in this CNN. The input into the model was a multidimensional tensor and a flattening layer was needed to reshape the array into a one-dimensional array that is readable for the dense layer and thus the output of the coordinates. A dropout layer was then used to prevent overfitting and enhance performance on the test images. This layer randomly removes inputs during training. It is a form of regularization that can be used, so that the model does not overfit the training data. The dense layer forms a linear connection between nodes and prepares the network to produce an output of four numeric values, which are the coordinates of the predicted bounding box. A dense layer of 256 units and then 4 units were used to complete the model. The final layer has 4 units because each observation will have an output of 4 coordinates.
Model Training
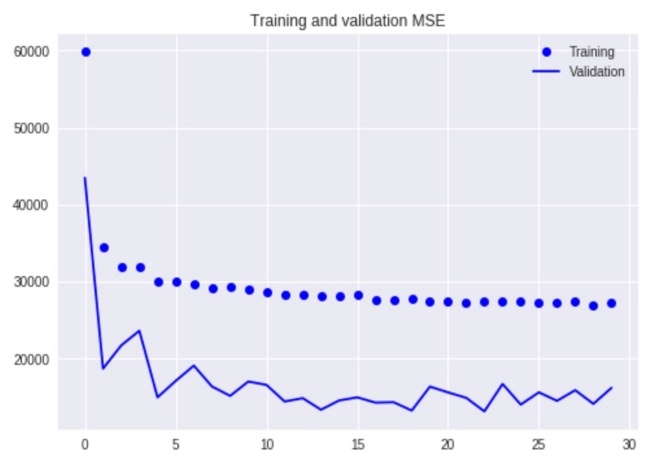
The initial results from the CNN showed evidence that the model was learning as the number of epochs increase, however, I found it puzzling that the training error was higher than the validation. The training error is expected to have lower error than the validation error because the model is being test on the validation set and was not used when adjusting the model’s weights. It is possible that there was evidence of data leakage, in which there is an overlap in the images used between the two datasets.
Model Evaluation
Bounding box models are typically evaluated using a metric called Intersection Over Union (IoU) and it is computed using the coordinates of the true bounding box and the predicted one. IoU is the ratio between the overlapped area between the two boxes and the total area that is not overlapping. This gives a metric to evaluate whether the model predicted a box that is close in proximity to the actual box.

An IoU score of 0.5 is commonly considered to be the threshold for a “good” prediction as Adrian Rosebrock, PhD explains here. The mean IoU score from the model was .52, so it’s a relatively well-performing mode in terms of average prediction.

Although the average prediction was greater than the .5 threshold, many predictions seem to be biased due to some images having multiple scans, like the one below which had the lowest IoU score of 0.111.
Limitations

The image above has two scans and thus, two bounding boxes must be predicted. The dataframe is organized so that each image has one set of box coordinates, and if an image has two lesions, the same image is listed twice each with different coordinates. This is problematic because when the model attempts to locate the lesion, there are two possible answers, which introduces unnecessary complexities to the model. One way to lower the error further would be to vertically split the “double” images in half, so that each image only has one true bounding box.
Another problem with the predictions is that the size of the bounding box seems to be inaccurately scaled as seen in the below picture, which had the highest IoU score of .882. This reveals an inaccuracy in the actual predicted bounding box, which gives strong evidence that the coordinates were not scale correctly when resizing the image to a 128 x 128 dimension size. There is great overlap between the two, but unfortunately, the true values are not correct.

Another possibility for future research would be to normalize the coordinates into a range between 0 and 1. This would allow the final layer of the CNN to have a sigmoid activation, which would most likely yield more accurate coordinate predictions. An additional change that may improve results would be to transform the predicted values to be the coordinates of the box’s top-left corner along with the box’s width and height, instead of the coordinates for opposite corners. This is only a subtle difference, but it worth experimenting with.

