The Great Escape: Health, Wealth, and the Origins of Inequality
Contributed by David Comfort. He took NYC Data Science Academy 12 week full time Data Science Bootcamp program between Sept 23 to Dec 18, 2015. The post was based on his second class project(due at 4th week of the program).
Overall Goal: Build a companion interactive presentation for a book, The Great Escape: Health, Wealth, and the Origins of Inequality, by Nobel Prize-winning economist, Angus Deaton.
Specific Goal: Build an interactive presentation with embedded Shiny apps
Tools Used: RStudio, Google Charts and GoogleVis
Angus Deaton, winner of the 2015 Nobel Prize in economics, is a Professor of Economics and International Affairs in the Woodrow Wilson School of Public and International Affairs and the Economics Department at Princeton University.
In The Great Escape, Angus Deaton–one of the foremost experts on economic development and on poverty–tells the remarkable story of how, beginning 250 years ago, some parts of the world experienced sustained progress, opening up gaps and setting the stage for today's disproportionately unequal world.
The interactive presentation with the Shiny apps embedded in it, is live at http://216.230.228.88:3838/bootcamp003_student/Project2-Shiny/DavidComfort-Shiny-Project/Great-Escape-v3.Rmd.
A screen capture of the presentation is below:
[youtube http://www.youtube.com/watch?v=qElqGyCSB9k?rel=0&w=560&h=315]
Outline of creating an interactive presentation with embedded Shiny apps
- Create a new R Markdown file in R Studio.
- Select Presentation and HTML (ioslides) when creating the presentation (see Figure 2.)
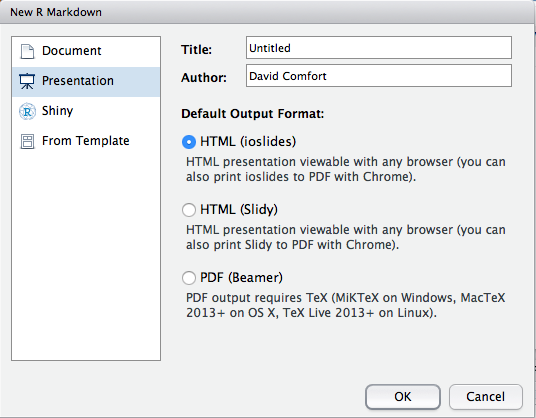
Figure 2: Create an interactive presentation in R Studio
- The header of the file now contains the following:
- Now, the
ioslides_presentationoutput format is specified in the front-matter of the document. - We have added our own CSS to the ioslides presentation using the
cssoption. The subdirectory where the css file,style.cssis located is specified as well bywww. - When knitr processes an R Markdown input file it creates a markdown (md) file which is subsequently tranformed into HTML by pandoc. If you want to keep a copy of the markdown file after rendering you can do so using the
keep_mdoption. - You can add a logo to the presentation using the
logooption. - You can customize the speed of slide transitions using
transitionoption. This can be “default”, “slower”, “faster”, or a numeric value with a number of seconds (e.g. 0.5) . - You can display the presentation using a wider form factor using the
widescreenoption. - The front matter also contains
runtime: shiny, so you can embed shiny apps into the document. Once you’ve addedruntime: shinyto the document you can run it using either the Run Document command in RStudio or using thermarkdown::runfunction. By default, documents are re-rendered on save, so once you’ve got a browser open with the document loaded, just save the R Markdown file to see your changes. - You can create a slide show broken up into sections by using the
#and##heading tags (you can also create a new slide without a header using a horizontal rule (----). For example here is the beginning of our slide show:
- Embed a Shiny app within one of the slides by defining the application inline using the
shinyAppfunction:
- Note that in all of R code chunks above the
echo = FALSEattribute is used. This is to prevent the R code within the chunk from rendering in the document alongside the Shiny components.
Specific Steps for embedding our Shiny App
I embedded several GoogleVis Motion Charts in my interactive presentation. I went over how I created a GoogleVis motion chart using R and R Markdown in a previous blog post, Gapminder Data Visualization using GoogleVis and R.
The specific steps involved were:
- Load
googleVisandshinylibraries - Specific the input choices,
myChoices, for the checkboxes - Load the
rdafile (previously saved) into an R data frame,gapdata - Set the
Sub.Regioncolumn of the data frame as a factor
- Call the
shinyApp. - Set up the
ui. - I have used a
fluidPageand put the interactive element, the checkboxes, in a sidebar. - Note that the name of the element is "Sub.Region", the elements are the elements specified
myChoicesabove. - I have also chosen to have have all elements checked using selected =
myChoices. - The main panel simply has a placeholder for the output of the motionchart, called
motionchartfor simplicity.
- The next section calls the shiny server.
- On the server side, Shiny applications use the
inputobject to receive user input from the client web browser. The values ininputare set by UI objects on the client web page. - There are also non-input values (in the sense that the user doesn’t enter these values through UI components) that are stored in an object called
session$clientData. - To access
session$clientDatavalues, you need to pass a function toshinyServer()that takessessionas an argument (sessionis a special object that is used for finer control over a user’s app session). Once it’s in there, you can accesssession$clientDatajust as you wouldinput. - The
target_gapdata"listens" to the client as far as which checkboxes are selected and creates a subset of the data frame based upon whichSub.Regionsa user has selected. It is important that this statement resides outside the output statement below. - Then, the output$motionchart statement calls
renderGvis. - The statement,
target_gapdata <- target_gapdata(), calls the aforementioned statement to select only the rows for theSub.Regionswhich have been selected by the user. - A Google Motion Chart is returned.
- The parameters for
GoogleVisinclude:data: a data.frameidvar: the id variable , “Country” in our case.timevar: the time variable for the plot, “Years” in our case.xvar: column name of a numerical vector in data to be plotted on the x-axis.yvar: column name of a numerical vector in data to be plotted on the y-axis.colorvar: column name of data that identifies bubbles in the same series. We will use “Region” in our case.sizevar– values in this column are mapped to actual pixel values using the sizeAxis option. We will use this for “Population”.
- The
options=list(state=statement is used to specify the initial conditions for the chart. This state can be copied from an existing chart by going to advanced settings and copying the state string. (See Figure 3.)

Figure 3: Setting State for GoogleVis
- See more information about how to create a GoogleVis chart using R at: http://nycdatascience.edu/blog/students-work/gapminder-data-visualization-using-googlevis-and-r/.
Embedding Images, articles and Videos in an interactive presentation
- Images can easily be embedded in the presentation by using the following format:
<img src="www/equation.gif" alt="Happy Planet Index"></img>
- It is also possible to embed videos by simply embedding an iframe with the following format:
[youtube http://www.youtube.com/watch?v=XwLNqDbPNBw?rel=0&controls=0&showinfo=0&w=853&h=480]
An article can also be embedded using an iframe:
Lessons Learned
- I had tried to create an interactive presentation using a package, Slidify, but I could not get it to work with GoogleVis. So rather forcing a tool, Slidify, to do what you want it to (and ends up not working), find another tool, Rstudio ioslides (that works) to do the project.
- A lot of well-known scientific papers and books are not easily reproducible and a lot of data is not publicly available, or at least readily available.
- A lot of data on important issues is simply missing, especially going back in time.
- It would be great to have companion websites / applications for papers and books which rely upon data analysis and visualization.
The interactive presentation with the Shiny apps embedded in it, is live at http://216.230.228.88:3838/bootcamp003_student/Project2-Shiny/DavidComfort-Shiny-Project/Great-Escape-v3.Rmd.
More information about creating interactive presentations using R, R Markdown, Shiny and Knitr can be found at:

