Semantic Segmentation of Nodules in Thyroid Ultrasound Images Using a Fully Convolutional Neural Network
Introduction
Complications in biopsies can lead to hemorrhaging, infection, and damage to nearby tissue or organs. To reduce the frequency of inaccurate readings and false positives resulting in unnecessary biopsies, Koios Medical uses image analysis and artificial intelligence algorithms in the early detection and treatment of disease.
In collaboration with Koios Medical, the goal of this project is to perform semantic segmentation of nodules found in thyroid ultrasound images. Semantic segmentation is the process of associating each pixel in an image with a class label. For this project, a pixel is either labeled as nodule or non-nodule.
The Digital Database of Thyroid Ultrasound Images is an open source database that contains 345 patient cases and 635 images with coordinate locations of nodules. A fully convolutional neural network referred to as U-net is constructed in Python using Keras and Tensorflow backend for fast and precise image segmentation of nodules.
Data Pre-Processing
All images have associated XML files containing metadata about the image. A dictionary data structure is created using image id as a key to map all coordinates of individual nodules denoted as marks to the associated image. A data frame is constructed with the dictionary and used to overlay coordinates onto the associated images for spot checking. The figure below illustrates coordinates overlayed on an original image.
A separate image is created with coordinates only. The coordinates are used to create a contour. The image is then binarized by setting pixels within the contour to 255 and all remaining pixels are set to zero. The newly constructed image is a mask used as a label for model training. The mask for the image above is shown below.
Nueral Network Construction
The U-Net is a convolutional neural network that was developed for biomedical image segmentation at the Computer Science Department of the University of Freiburg, Germany. The network is based on the fully convolutional network and its architecture was modified and extended to work with fewer training images and to yield more precise segmentations. The architecture is illustrated below.
Below is a summary of the modified architecture used. Training was performed with a various number of filters, with and without batch normalization, with and without dropout, and with and without data augmentation.
Due to the small sample of ultrasound images, data augmentation is used to create an additional 1500 images. A series of random rotations between ± 20 degrees and horizontal flips are performed. Random augmentation is applied simultaneously to image and mask label to maintain a relationship. A sample of an augmented image and assocated mask is shown below.
Results
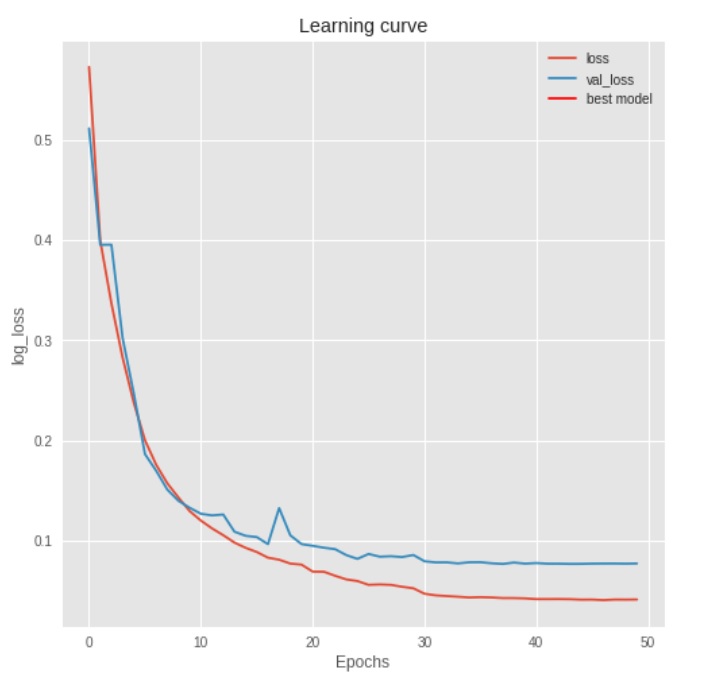
The best results are achieved using data augmentation with batch normalization and without dropout. The best model had a mean Intersection Over Union (IOU) of ~.86 on the test set.
The binary cross entropy loss function is continually minimized until 30 epochs. This achieved a mean IOU of ~.86. Below are five random test set samples. The leftmost plot is the original ultrasound image with coordinate annotation. The middle left plot is the mask created from the annotations. The middle right plot is the predicted mask as determined from the trained neural network. The rightmost plot is the binarized prediction from the neural network. The associated IOU is found in the upper left corner of each image set.
Future Work
Further exploration into creating an end-to-end function that does segmentation and classification creating an output folder with cropped images in order of their probability of malignancy is the next stage.
All data and code can be found:
https://github.com/Joseph-C-Fritch/unet_segmentation