The GC Skew - Studying Genome Evolution
The skills the author demoed here can be learned through taking Data Science with Machine Learning bootcamp with NYC Data Science Academy.
Significance of the GC Skew
It has long been known that the four nucleotides are not equally abundant in bacterial genomes. Specifically, guanine is more abundant than cytosine and adenine is more abundant than thymidine along the leading strand of the replication fork. Over any given region, this difference can be expressed as either (G-C)/(G+C), or (A-T)/(A+T) commonly referred to as the AT and GC skews, respectively. Nucleotide skews such as the GC skew can be used as a tool to identify the replication origin and terminus of the chromosome.
When a typical bacterial replication origin fires, two replisomes initiate and move in opposing directions. Due to the anti-parallel nature of DNA strands, the leading strand of the two replication forks copy opposing strands. This asymmetry, and the accompanying mutational footprint, appear to drive opposing GC skew values across replication terminus and origin sites. The terminus and origin regions can therefore be inferred from the chromosome sequence of a new bacterial isolate, even if no other information is available. The DoriC database offers estimates for the location of both sites for thousands of bacterial species.

GC Skew as a tool for identifying recombination events - Pt. 1
The GC skew may have other uses as well. The mutational signature of the replication fork leaves an imprint on (potentially) every gene vis-à-vis the positive GC skew. This makes it possible to identify each gene's typical orientation. And if a gene undergoes a genomic rearrangement causing it to be encoded on the opposing strand, its GC skew value will concomitantly invert (negative to positive or positive to negative). This is because physical inversions cause every G in the top strand to be replaced by a C, and every C to be replaced by a G. Therefore, our group hypothesized that local GC skew inversions (areas with negative values) may be caused by a physical DNA inversion.
GC Skew as a tool for identifying recombination events - Pt. 2

Figure 2. Inversions in the GC skew suggest the presence of a local fragment inversion. (Adapted from Merrikh et al., Genome Biology and Evolution, 2018, Figure 1.). Gene locations are depicted in red/black on the outter ring. GC skew values are indicated (green/purple) on the inner ring. Gray arrows indicate local GC skew inversion regions.
In a recent publication, we performed a systematic investigation of GC skew values genes on several bacterial genomes. This led us to observe that most negative GC skew genes are encoded on the lagging strand. As such, we inferred that most lagging strand genes are actually native to the leading strand.
The idea that lagging strand genes were created through inversion is particularly interesting because expression of these genes can cause major problems for the cell: transcription of lagging strand genes causes RNA polymerase molecules to translocate in the direction opposing the movement of the replication fork. This can result in severe head-on encounters between the two machineries, interrupting replication and often breaking the DNA strands.
We (and others) have a long-standing hypothesis that an ideal cell might have all of its genes encoded on the leading strand, minimizing DNA replication stress. Our GC skew data offers some support for this notion, implying that most of the genes on the lagging strand are actually native to the leading strand.
However, our interpretation of GC skew values is somewhat contentious. Others suggest that the GC skew can become negative through mechanisms other than gene inversion. If correct, this would certainly undermine our interpretation of negative GC skew regions.
GC Skew as a tool for identifying recombination events - Pt. 3
To test the competing hypotheses, we examined the GC skew of all genes using two different calculations. Specifically, we calculated the GC skew of nucleotides that should be able to mutate quickly (codon position 3 nucleotides) versus those that cannot mutate easily (codon position 1 nucleotides are constrained because nearly every nucleotide change will result in a change in the encoded amino acid, thereby altering the function of the encoded protein).
If the competing hypothesis is correct, the codon position 3 nucleotides should have a lower GC skew than the position 1 nucleotides. (i.e. indicating that mutational pressure drives increasingly negative GC skew values in many lagging strand genes.) Conversely, if our hypothesis is correct, easily mutable nucleotides (position 3) should have the same positive GC skew apparent along the rest of the chromosome. This should be true even for lagging strand genes.
While an inversion would initially cause the GC skew of both position 1 and position 3 to be negative, under our model, the mutational pressure should drive the value to become increasingly positive and this would have a more potent effect on position 3 nucleotides. It also sets up the expectation that position 1 nucleotides should retain the negative GC skew. To assess this hypothesis, we applied the two methods to every gene in several bacterial genomes and binned the values by organism and gene orientation using the GC skew plotter R Shiny app:

GC Skew as a tool for identifying recombination events - Pt. 4

Figure 3. GC Skew value distributions for leading and lagging strand genes in M. tuberculosis. GC skew values based upon codon position 1 (upper graph) or position 3 (lower graph) are depicted for leading strand genes (CD/Codirectional, Red) or lagging strand genes (HO/Head-On, Blue). If negative GC skew values are indeed due to inversions, codon position 1 based GC skew values should be negative while the same genes should have positive codon position 3-based values.
As you can see from the upper graph above (Fig. 3), codon position 1 based GC skew values have a bimodal distribution in M. tuberculosis whereas the position 3-based values have a single distribution. By binning the data by gene orientation, it became clear that negative GC skew values are almost entirely correlated with the lagging strand (HO) orientation (Fig. 3, red vs. blue, upper graph).
Additionally, the same lagging strand genes have a positive codon position 3-based value (Fig. 3, lower graph, blue) suggesting that mutational pressure drives the GC skew to become increasingly positive with respect to the leading strand (blue profiles in the upper versus lower graphs). These data are consistent with the idea that easily mutable nucleotides (codon position 3) were rapidly mutated due to the mutational signature of normal DNA replication, causing them to acquire the positive GC skew apparent in the rest of the strand. Hence the leading/lagging strand values are essentially the same for position 3.
Meanwhile, the less mutable nucleotides (codon position 1) retained the highly negative values caused by the inversion. This pattern is even more striking for the Gram positive species B. subtilis. Notably, B. subtilis is in a completely different bacterial phylum, illustrating the consistency of this pattern across species. Additional data for E. coli and Mycoplasma gallisepticum can be examined here.
GC Skew as a tool for identifying recombination events - Pt. 5

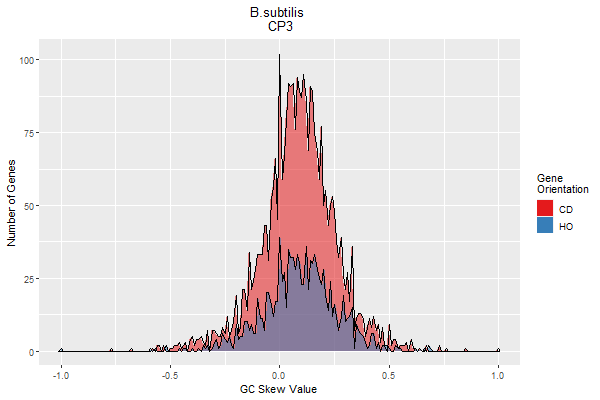

Figure 4. GC skew value distributions for B. subtilis genes. Graphs were created using the GCS PlotR app. Upper graphs depict the codon position-specific GC skew value distributions (Codon position 1 in the upper graph, and position 2 in the middle graph). The bottom graph depicts the same codon position 1 based values of individual genes in the form of a bar plot, binned by gene orientation. The values are sorted based upon the whole-gene GC skew value (not shown), resulting in the appearance of a curve. The high-to-low value distribution of these data shows that the codon position 1-based GC skew values mirror the whole-gene-based method. This shows that the position 1-based calculation is consistent with the typical GC skew calculation used to identify the chromosomal origin and terminus. Note that there are fewer lagging strand (HO) genes than leading strand (CD) genes, resulting in a smaller data set (blue).
In other words, gene orientation appears to be sufficient to explain the bimodal GC skew distribution (Z-test p-value = 0) and negative GC skew values. Additionally, as mutational pressure seems to drive an increasingly positive GC skew, it appears to be unlikely that mutational pressures cause the negative GC skew of many genes. Overall, these observations are consistent with the idea that negative GC skew values are an indication that a gene is in the inverted state (relative to its typical orientation). By extension, our findings suggests that most lagging strand genes were likely the result of a gene inversion event that brought a natively leading strand gene to the lagging strand.
Conclusions and Future Work
Overall, these data offer a variety of insights into the evolutionary history of the bacterial chromosome. They suggest that there is a significant benefit to having genes encoded on the leading strand, co-orienting transcription with the DNA replication machinery. However, in our original manuscript we provide additional analyses of negative GC skew genes and show that, at least temporarily, the lagging strand (HO) orientation may be beneficial to the cell. We provide additional evidence head-on replication-transcription conflicts confer a faster rate of evolution on many inverted genes, potentially providing this benefit.
We hypothesize that in some contexts, the increase in mutagenesis and subsequent faster evolution may be enough to offset the cost associated with the decrease in DNA replication speed.
While our work offers a tempting hypothesis about the evolutionary history of bacterial genomes, there are a variety of influences affecting the GC skew. These including sequence context and selection, as evidenced by the work of Chen et al. among others. While the data presented here implicates gene inversion as the predominant mechanism causing negative GC skew regions, this is only one interpretation of the data. Future work is needed to help determine if gene inversion is truly the only major cause of negative GC skew regions and define the relative effects of other influences.
GCS PlotR App
GC skew profiles were plotted using ggplot2 using the GCS PlotR Shiny app, available on GitHub. The app allows users to to visualize the GC skew value distributions by codon position, and gene orientation for four bacterial species: E. coli, M. tuberculosis, B. subtilis, and M. gallisepticum. The tool can also be rapidly customized to display equivalent data from other organisms.


