Predicting Hospital Readmission Rates
Introduction:
In this project we set out to train a machine learning model that would flag patients from a skilled nursing home if they are at risk of being readmitted to the hospital. Preventing patient readmission to hospital can improve the patient experience, lower costs, and, hopefully, lead to a shorter stay in the skilled nursing home. We were given two datasets from ~10 different nursing homes in NYC that specialize in patients with acute/chronic heart failure. We set out to merge the two datasets and combine various metrics (ie. lab results, patient’s weight) to have one unique observation per patient with all of their vital statistics and their outcomes (Home or Readmission to Hospital). We used and compared a logistic regression model and a tree-based XGBoost model to predict outcomes. We used Google API to create a pipeline that will pull all the data and update our predictions and evaluation of our model.
Cleaning the dataset:
The number of patients in the dataset was limited, which was a challenge given that the more observations you have, the better your predictive model is likely to be. As our goal was to predict whether patients will be discharged to a hospital, we treated each period from enrollment in the program to discharge for any patient as a unique observation within out data set. This means that multiple observations can be derived from the same patient if they have been through the program multiple times, increasing the number of observations we could use to train our model.
Unfortunately, data for each observation was plagued with missing data, extreme outliers, and nonsensical values, like negative ages for example. Fortunately, some of the features had redundant entries within the spreadsheets. So if a value didn’t make sense, we could use the value from another part of the spreadsheet. Sometimes, values were entered into the spreadsheet in different formats, a mixture of percentages and decimals, for example. In cases like that, it was clear how to fix the values. Any nonsensical values that couldn’t be fixed were treated as missing values.
After removing observations with too much missing and erroneous data, we had fewer than 500 observations with which to train our model.
Features in the dataset:
Within the dataset, there were 31 variables of interest. They consisted of the patient’s age, gender, blood pressure, ejection fraction, length of stay, diagnoses, weight, medications, and various lab results. Since a patient could have multiple diagnoses, the diagnosis variable was parsed out and dummified into 8 unique diagnosis variables (cad/mi, heart failure unspecified, diastolic heart failure, systolic chronic heart failure, atrial fibrillation, cardiomyopathy, left ventricle assisted device, chronic heart failure).
Here we have a first look at all the variables of interest lined up in a correlation matrix. Lighter values indicate positive correlation, and darker values indicate negative correlation. Based on this, there does not seem to be any strong correlation between the predictors and our outcome variable. Predictors that show correlation amongst themselves are BUN & CR, BUN & BNP, CR & BNP, and systolic & diastolic. The first three pairs are related to kidney function. and the last is related to blood pressure.
One variable that we expected to have a greater impact on our outcome variable was EF. After graphing the distribution of EF by outcome (below), we see that the distributions are fairly similar.
Regarding the distribution of weight change as a fraction by outcome (below), there is slightly higher positive weight change in patients with a negative outcome. Overall the distributions are similar and echo what is shown in the correlation matrix.
For medications, those with positive outcome were seen to have a slightly higher frequency of prescriptions (except inotropes), but overall the distributions are comparable.
For the eight unique diagnoses, we noticed ‘lvad’ and ‘chf’ have low values and, as such, may not be important variables to keep as predictors. Further domain knowledge would help us better understand whether or not to drop these variables.
Missingness imputation:
In any real-world data analysis, an unavoidable hazard is missing values. Our dataset also had some features with many missing values.
Machine learning algorithms often have difficulty dealing with missing values. Nevertheless, it’s not ideal to remove all observations with missing values, as it can severely reduce the size of data or the data on one particular feature. Not all missingness is the same. There are different kinds that require different forms of handline. If the probability of missingness in a feature is the same for all events, we call it missing completely at random. When the probability of a variable being missing depends only on known features, the missingness is said to be random. If data are missing completely at random or at random, it is acceptable to drop events with missingness as it will not distort our distribution.Some of the missing values in our dataset is due to error in data input. It is reasonable to assume these are missing completely at random and safe to drop without causing significant distortion to the distribution.
However, if the missingness is not random, dropping events may distort the dataset. For example, if in our dataset we observed many missing values for blood pressure. When we found that patients with low blood pressure did not have their blood pressure reported, then that will be an example of missingness not at random. Dropping observations with missing blood pressure values in that case will bias our distribution, and any model built on that data will likely be flawed. We assumed that missingness in our dataset is completely at random or at random.
In most cases, instead of dropping observations with missing values, we Imputed values for features using different techniques. For example, missingness in weight change or blood potassium level was imputed by median of the known values for those variables, whereas missingness in “acute or chronic” state of patients was imputed by the duration of stay in the nursing home. Some of the patients were missing data in the gender column, so we filled in the missing data using a function called Gender Guesser. This function takes the name of the patient from the name column and decides whether it is a male or female. This made it possible for us to retain many observations. It should, however, be noted that imputing missing values in this way may underestimate the standard error.
Feature scaling:
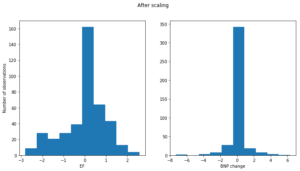
We also decided to employ feature scaling to some of the numerical features in our dataset. We subtracted the mean of the variable from individual observation and scaled it to unit variance. Standardization prevents the cost function from being dominated by features with a large range. It also makes gradient descent converge much faster to the minima of the cost function. For example, the ejection fraction and bnp change variables before scaling have drastically different ranges.
After scaling the two distributions have comparable range (along the x-axis).
Feature transformation:
Some of the numerical features in the dataset had skewed distribution. Taking the logarithm of a skewed variable may improve the fit in the modeling stage by making the variable more "normally" distributed.
PCA and Logistic regression:
After all the cleaning, feature selection and engineering was done, we had over forty predictors in our model. Trying to understand and visualize data with over forty dimensions is, needless to say, difficult. So we used principal component analysis to reduce the dimension of our data down to a much more manageable three dimensions. Here is the scatter plot of the observations in our data set. Good and bad outcomes are colored blue and red respectively.
http://plot.ly/~SimonJoyce/0.embed
Unfortunately, we can see that the outcomes are mostly mixed uniformly in the data set. This meant that most of the more classical prediction models, such as logistic regression and discriminant analysis should perform poorly. Nevertheless, we tried a logistic regression with gridsearch and 5-fold cross validation. It performed fairly well, with a cross-validation score of 0.61 and a hold-out score of 0.68. We decided to also try and compare a tree-based model, using XGBoost.
XGBoost:
In order to better understand why this model was appropriate for the project, we first need to familiarize ourselves with two machine learning techniques: decision trees and boosting.
Decision trees are frequently used as estimators in machine learning. A decision tree uses a set of rules to predict an outcome based on one or more predictors, or features. Trees are popular because they are easy to implement and simple to interpret when represented graphically. To demonstrate this, let’s inspect the decision tree graphic below.
We have a collection of patients, and we would like to predict whether they will have a positive or negative outcome when leaving the hospital. A positive outcome is represented by 1, and a negative outcome is represented by 0. Each patient has a set of measurements associated with them (e.g. cad.mi, bun, bnp, duration, resting_hr, etc.). The decision tree makes predictions based on a set of rules it produces. These rules allow the tree to make decisions about how to organize patients into different groups and, ultimately, assign a prediction to each group of patients. In the tree above, the first decision is made based on a patient’s “cad.mi” value. Patients with “cad.mi” less than 0.5 follow the branch down to the left, whereas all other patients follow the branch down to the right. The patients continue to be split based on new rules. The point at which the tree stops splitting is called a terminal node.
In the decision tree above, the terminal nodes all have a value of either 0 or 1. The value at the terminal node is the predicted patient outcome. For example, if we start at the top of the tree and continue left at every node split (i.e. the patient satisfies the rule at each split), we will end up with a negative predicted outcome. Although in the tree above we see a 0 or a 1 at each terminal node, the actual value behind the scenes is a probability that the outcome will be positive. The probability can be useful if we want to set a threshold for considering a case positive (i.e. only groups with probability greater than 70% will be considered positive). The threshold is relevant when we want to limit the number of false positives by setting stricter criteria for predicting a positive outcome. This is particularly important in health care because false positives can turn into dangerous situations. If we raise the threshold, the model will predict a positive outcome only when we are sufficiently confident about it.
Now that we have an understanding of how decision trees work, we can discuss a powerful technique in machine learning called boosting. Boosting is a method that starts with a weak decision tree and improves its accuracy by sequentially adding corrections. Consider this example: my friend Sophie is decent at guessing someone’s age. I notice that whenever Sophie guesses an age, she is off by +2 years for men and -2 years for women. If we correct Sophie’s guess using the pattern of error I have observed, we end up with a more accurate prediction. We can continue to correct Sophie’s guess by finding trends in her error until we have a much more powerful model than we started with. This is the main idea behind boosting.
XGBoost is a very popular machine learning model that utilizes the concepts of decision trees and boosting. XGBoost stands for extreme gradient boosting and it does a great job of pumping the most predictive power out of a decision tree based model. It is a powerful and efficient algorithm for large datasets. We test this model in an attempt to pull the most predictive power out of our dataset. Not surprisingly, it performs better than a simple logistic regression: the model had a cross-validation score of 0.66 and a hold-out score of 0.69. However, with such a small dataset, XGBoost’s optimization for speed is not as important to us as the explanatory power of a linear model like the logistic regression.
Model evaluation:
In order to compare our models, we plotted the ROC curve (Receiver Operating Characteristic Curve), which compares the True Positive Rate and the True Negative Rate at varying thresholds for classification. This is a way to visualize the performance of a model without narrowing down to accuracy, precision, or recall, for example. An ideal ROC curve will be as close to a right angle as possible. With XGBoost, the terminal nodes each have a probability of being in one or the other class (Home or Readmitted to Hospital). With logistic regression, the output predictions all fall on the sigmoid function between 0 and 1; they are also probabilities. In order to compare these results with the actual outcome from our training set, we can change the threshold to be in one class from the default 50% to something more strict or flexible. In this case, we would want to maximize our True Negative Rate and minimize any False Positives, which would be patients we classified as Home who are actually not doing well. See the figures below which show the ROC curves for both our models. The ROC curve allows us to evaluate our model regardless of the threshold we choose and to see the trade-off between specificity and sensitivity.
The first thing we can note is that the AUC score of the logistic regression does not change much from the training set to the test set, while the XGBoost performs too well on the training set. The huge decrease from 100% to 79.6% on the test set is a sign of overfitting. Because we are dealing with a relatively small data set, we want to be cautious of overfitting, as this could lead to very inaccurate predictions on future data. If we change the hold-out test set in this dataset, the XGBoost might have a very different area under the curve, while the logistic regression would stay relatively stable. Therefore, despite the higher accuracy of the XGBoost model, we prefer logistic regression for its stability.
Deployment:
Since our collaborating partners used a front end AppSheets, with Google Sheets as the backend, we used Google API to be able to download the dataset, clean, make our predictions, and upload our predictions to a specific column of the Google Sheets. This would allow certain patients to be flagged in the front end, based on the value of that column in the back end. We also added a tab to the Google Spreadsheet that would have information about the model’s accuracy, the last time it was run, and a history of its accuracy over time. We would hope that with more observations, our model would become more accurate.
Future Work:
We were very excited to be able to work with a real-world healthcare dataset for this project. Our goal with this project was to start with the simplest model possible, and if we had time, expand from there. Within the scope of two weeks, that means we chose the most recent measurements for each patient. For example, it made no difference if a patient had been weighed once or twenty times during their time in the nursing home; we only evaluated the most recent weight. Since most of these measurements were taken twice a month, this was overlooking a vast dataset behind the scenes, albeit a complicated one. One way to simplify it would be to treat each week a patient is in the hospital as an observation, with new measurements or not, and have a multi-class outcome: Home, Stay, and Hospital. While complicating the model significantly, this would lead to a much more powerful and useful model. Predictive analytics is vastly changing our healthcare system in lowering costs and improving patient experiences, and we were excited to work in this area. Please feel free to reach out with any additional questions or comments on our work.
Summary of Results
| Logistic Regression | XGBoost | |
| Cross Validation Accuracy | 61.1% | 65.69% |
| Train Accuracy | 69.7% | 99.6% |
| Test Accuracy | 67.92% | 68.69% |
| Threshold for Classification | 49.18% | 83.9% |
| Test Accuracy (with threshold change | 68.87% | 59.6% |
| Precision (with threshold change) | 66.07% | 82.35% |