Redefining Cancer Treatment: Predicting Gene Mutations to Advance Personalized Medicine
Introduction and Project Scope
One of the most exciting frontiers for machine learning is the field of medical diagnosis, where background information and nuanced decision-making are considered essential. When Memorial Sloan Kettering (MSK) released a Kaggle competition entitled “Personalized Medicine: Redefining Cancer Treatment,” we took on the controversial challenge of creating models that could perform the same tasks as experienced genomics researchers. We were provided with a dataset of genes and cancer-related mutations, a text file of scientific literature related to each mutation, and a 1-9 classification of each case, which had been hand-annotated by a scientist at MSK after reviewing the data. Although we were not told in advance what each class represented, our goal was to correctly classify new “test” mutations after training machine learning models on the original datasets and text files; we were informed that our work would ultimately help automate the work of distinguishing between driver (cancer-causing) and passenger (neutral) mutations.
Recognizing that machine learning methods are still far from perfectly replicating the work of skilled oncologists, we put together a number of tools that cancer researchers can use in conjunction with the classification model to speed up this time-consuming process.
EDA and Feature Engineering
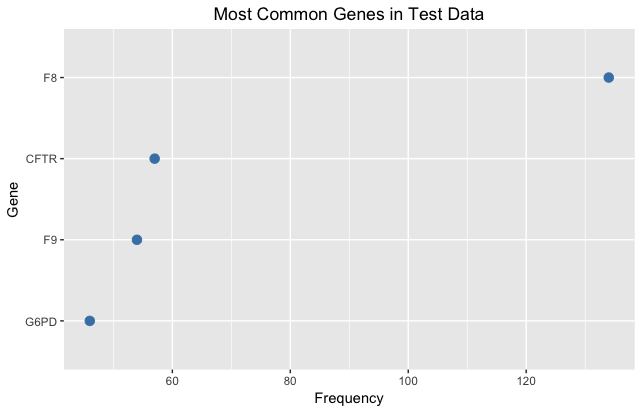
Our first step was to visualize the data and extract as much information as possible. We examined the relationships between genes and classes and found that one gene could fall into many different classes, suggesting that mutations within the same gene could produce widely different effects. We also noted that the majority of mutations in each case were point mutations, in which one amino acid was mutated to another. For these cases, we created two new columns in our data for the original and final amino acids and then merged our data with another dataset on the physicochemical characteristics of each amino acid. Using scales of amino acid charge and hydrophobicity (relationship with water), we created new features measuring the absolute value change in charge and hydrophobicity caused by each amino acid mutation; we reasoned that if the new amino acids had very different properties from the original ones, it was likely to lead to changes in overall protein structure and function. Though our feature engineering of the original dataset was helpful, we noted that the genes included in the training and test datasets were almost entirely different, and since the gene/mutation datasets contained limited information, we turned most of our attention to the text data.
Text Analysis and Vectorization
Our approach to the text data was two-fold. We aimed to 1) use our background knowledge of the scientific literature to create intelligent stopwords and filter the data meaningfully and 2) explore different natural language processing vectorization methods (n_grams, TF-IDF, Word2Vec, Doc2Vec) to translate the text into numeric features that our classifier can work with. Since we were dealing with large blocks of scientific text, our first goal was to extract only the relevant sections of text (e.g. include results, remove methods and materials). We also selected sentences containing words that were likely to contain other words that would help in classification, such as “passenger,” “driver,” “tumor,” etc. We found that using these informed subsets of the text worked well - our models using them often outperformed models containing the full text data.
Below: Visualization of differing word frequencies across classes
- TF-IDF
The first text vectorization technique we tried was TF-IDF, which rewards words that appear more often in a text, but penalizes words that appear in all texts, to help filter out common medical terms. We also manipulated parameters such as n_grams, which determined the lengths of grouped words that our model used.
- Word2Vec / Doc2Vec
Word2Vec is a word embedding technique that was developed by Mikolov et al. in 2013 at Google. The model itself is a shallow neural network with three layers: an input, output, and a hidden layer in between. Upon initialization, a word weight matrix is created, with a vector for each word in the corpus. The length of these vectors is a hyperparameter of the model, with the number typically ranging from 100 to 500. The values for this matrix are randomized upon creation. The inputs that are fed into the network are one-hot encoded vectors of each word (called the center word), and the output for each is the probability that all the other words lie in the context of the center word. This context is a sliding window around the center, and is another hyperparameter of the model. During each epoch of training, these inputs are multiplied by the weight matrix. The weights are then adjusted by backpropagation through stochastic gradient descent, so that the probabilities of the next epoch are closer to the true probabilities of the context. This weight matrix is what is used as the word embeddings in our model. There are two different training algorithms that Word2Vec can employ - continuous bag of words, or the skip-gram. The latter is better at predicting infrequent words, due to the fact that there is no averaging of the word embedding vectors during prediction time.
Doc2Vec is almost the same as Word2Vec, the biggest difference being the addition of a paragraph matrix. In Doc2Vec, the corpus is split into paragraphs, or, documents. This allows for more robust context inference, since each paragraph may have different contexts for each word. The paragraph matrix serves as a memory for these contexts. For each center word present in each paragraph, the paragraph vector is passed along with the word vector, and the weights are calculated for it as well. Doc2Vec also has two training algorithms that function is about the same way. The skip-gram algorithm (accessible through the ‘Distributed Bag-of-Words’ model) was used for out Doc2Vec embeddings.
Machine Learning Models
Our approach to the machine learning aspect of this project was to try as many classification techniques and feature combinations as possible - in hopes that each method is likely to bring a new perspective in capturing features unique to each class. Here we discuss the benefits and drawbacks to each technique generally, and how each performed in classifying mutations into MSK’s nine categories.
- Parametric
-
- Multinomial Logistic Regression (MLR)
Easy to interpret and popular across a number of industries, logistic regression fits a linear equation of variables to predict log odds, which is then piped through the sigmoid function to find the conditional class probabilities for each observation. Our best MLR model used 400 Doc2Vec features, 20 gene and 20 variation SVD features, the absolute value change in charge, and hydrophobicity. 5-fold cross validation led us to choose C = .1, L1 regularization, and balanced class weights to prevent the model from overpredicting more popular classes. Given the sensitivity of classifying mutations and their effect on cancer tumor growth, we decided that balancing class weights in our algorithms was essential, even if our prediction accuracy dropped. This particular MLR model achieved 59.5% classification accuracy and was fairly good at predicting all classes (e.g. it did not overpredict popular classes).
-
- Multinomial Naive Bayes
Multinomial Naive Bayes is often cited as a fast, effective text classification technique. In our case, given the relative word frequencies or other vectorized text features seen in research papers associated with each class, the model can predict the conditional probability of each class given a new observation’s word frequencies/text features. Rennie et. al. found that using TF-IDF or other text vectorization methods (rather than bag of words), as well as balanced class weights, helped improve MNB’s performance on text classification. These alterations helped MNB models overcome some classical difficult-to-attain assumptions, like as feature independence. We followed their lead. Our best MNB model used Doc2Vec and 20 gene/20 variation SVD features. We used scikitlearn’s MinMaxScaler to scale all features between 0 and 1, as scikitlearn’s MNB classifier cannot handle negative inputs. The best MNB model had an unimpressive 48.6% classification accuracy, overpredicting the most popular classes and underpredicting the less popular ones.
-
- Neural Network
Two deep neural networks were created for classification: one fully connected network, and one convolutional network. All networks were constructed using the Keras package.
The fully connected network consists of seven layers, the first five of which had 512 neurons, the penultimate had 100 neurons, and the output layer had 9. This architecture was devised through some cross-validation testing in terms of how deep (number of layers) and wide (number of neurons) the network was to be. The activation function for all layers but the output was the popular rectified linear unit, or ‘ReLU’; the output layer has a softmax activation function, since the output had to be the probabilities of the nine possible classifications. There were dropouts in between the layer, which remove a portion of the inputs in order to reduce overfitting. This network performed well, with an accuracy of 63.2%.
The convolutional network was modeled after a network used in a recent paper titled ‘Medical Text Classification Using Convolutional Networks’ by Hughes et al. (2017). It consists of two sets of two convolutional layers with a max pooling layer after each set. There is then a fully connected layer, followed by the nine neuron classification output layer. Convolution words by reading the inputs with a sliding window, so that all the fine details of the vectors can be read. The max pooling layers shrink the inputs, so that the memory use isn’t as extreme (convolutional networks require a lot of memory). This network performed slightly worse than the fully-connected network, with an accuracy of 60.1%.
- Non-parametric
-
- Support Vector Classifier
The Support Vector Classifier (SVC) is part of the larger model class of Support Vector Machines (SVM), which work by representing observations as points in space and then drawing boundaries between them that provide the clearest separations possible. SVCs work well for text classification since they can handle dense concepts and sparse instances. In problems involving text classification, models have to deal with many features, most of which provide some information. This leads to a high dimensional input space with few completely irrelevant features. Overall, SVMs can learn well independent of the dimensionality of the feature space, making them a good candidate for text classification. For our SVC model, we used TF-IDF for text vectorization and selected an n-gram value of 3, meaning that groups of up to three words could be selected together. Our SVC model predicted correctly across nine classes with 64.3% accuracy, making it one of our top performers.
-
- Random Forest
The random forest method constructs an ensemble of independent decision trees, each using a sample of training observations through bootstrap aggregation, and selecting a feature for each node split using a random subsample of features. Once all trees are fitted, the algorithm averages the class prediction probabilities for each feature-value combination to deliver the final model predictions. Our best random forest model used TF-IDF vectorization on the subset of text chosen through informed stopwords and n-grams = 3 -- good enough for 56.9% accuracy.
-
- Gradient Boosted Classifier (Including XGBoost)
Gradient boosted models are ensembles of weak learning decision trees. Instead of averaging the predictions of hundreds of independent trees like in random forest models, boosting takes an iterative approach - build a tree, calculate the output of your objective function (incorporating training loss and regularization), and use this output as an input for the subsequent tree’s objective function. Boosting adjusts the weights associated with each split in a tree based on the error from the prior tree - slowly descending along the error gradient (gradient descent) until the algorithm can settle on a local minimum that optimizes your objective function. A “learning rate” hyperparameter lets you decide the rate of descent.
For more on gradient boosting, we love the XGBoost team’s description here.
Our best GBM model had a classification accuracy rate of 67.8% and used the 400 Doc2Vec features, 20 gene/20 variation SVD features, absolute value change in charge, and hydrophobicity. 5-fold grid search cross validation helped us tune hyperparameters, eventually landing with 600 trees, .01 learning rate, maximum depth of 15, and minimum samples per leaf of 5. Analytics Vidhya has a great tutorial on tuning hyperparameters here.
We also used XGBoost on the same features described above. XGBoost offers two primary advantages over scikitlearn’s gradient boosting classifier, namely 1) it runs faster, and 2) includes regularization at each step of the process to control for overfitting. Our best XGBoost model achieved 70.1% accuracy. 5-fold grid search cross validation led us to a .01 learning rate, 1000 trees, a maximum depth of 15, and L2 regularization. The boosted models were generally good at predicting all classes.
General Model Takeaways
- All models confused Class 2 for Class 7 and vice versa, as well as Class 1 for Class 4, and vice versa
- Changing ‘class_weights = balanced’ reduced accuracy, but fixed overprediction of popular classes (important given our task at hand)
- XGBoost - most effective ML technique
- Doc2Vec - most effective text vectorization technique
- Ensemble of 8 models - performed worse than XGBoost mode
- Stack of 5 models using multinomial logistic regression - performed worse than XGBoost model